-
EN




Integrated Discovery Multiomics
In systems biology, there is a close upstream-downstream relationship among the genome, transcriptome, proteome/peptidome, and metabolome. Integrated multi-omics analysis enables the investigation of underlying regulatory networks across multiple omics layers, especially when single-omics data alone are insufficient to comprehensively decipher complex physiological processes. This approach provides deeper insights into disease progression and the functional pathways of biomolecules within the body.

Product Consultation
Well-healthcare Technologies possesses the capability to integrate multiple omics analyses—including small-molecule metabolomics, lipidomics, proteomics/peptidomics, and spatial metabolomics—within a unified detection system. This ensures maximum consistency across multi-omics samples and addresses the challenges of spatial-temporal stability in omics research, delivering high-throughput and highly stable results. Additionally, the platform supports integrated analysis with data from non-mass spectrometry technologies, such as genomics, transcriptomics, microbiomics, and radiomics, enabling multi-modal data integration. This provides a broadly applicable strategy for comprehensive investigations into biological systems and disease mechanisms.
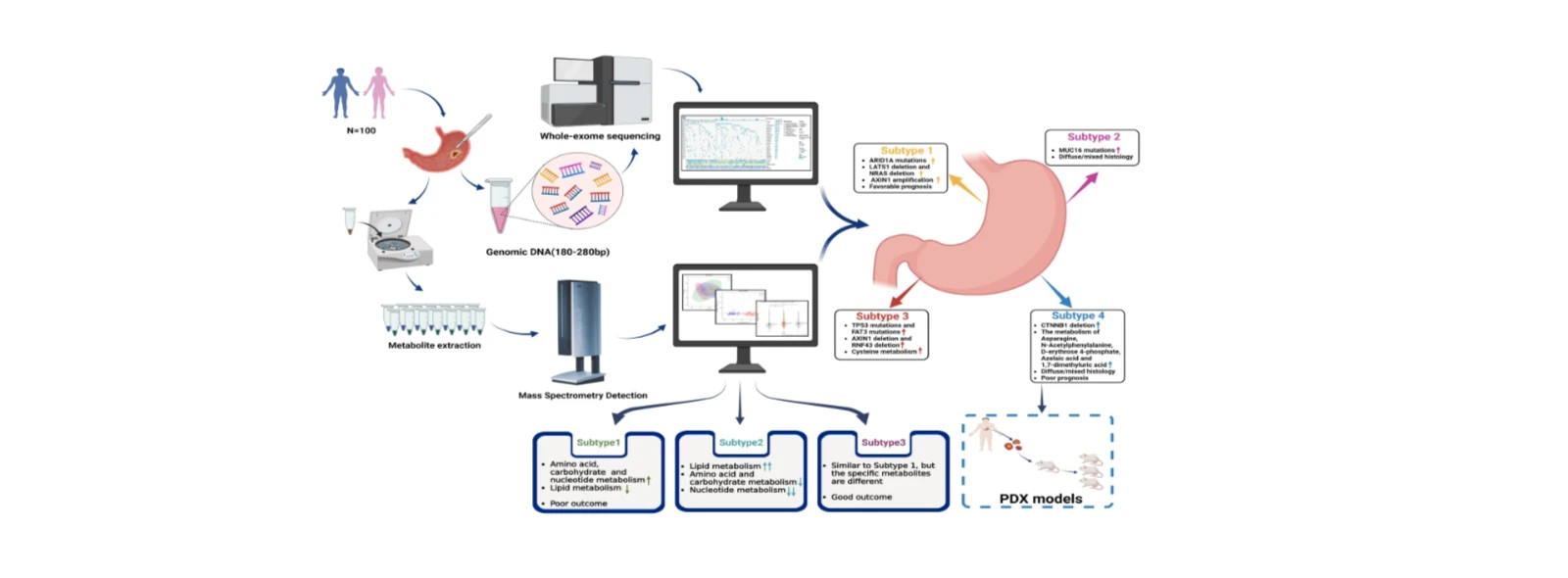
Establishing a Molecular Classification System for Gastric Cancer by Integrating Exome Sequencing and Metabolomics
Journal: Journal of Experimental & Clinical Cancer Research (JECCR), Published: February 2024, Impact Factor: 11.3
This study enrolled 100 Chinese gastric cancer patients with complete follow-up data and performed whole-exome sequencing (WES) and metabolomic profiling to characterize genomic mutations and metabolic alterations specific to this population. Based on metabolomics data, unsupervised clustering was used to identify metabolic subtypes, and differential metabolic pathways were compared across subtypes. Kaplan-Meier survival curves and regression analyses were employed to explore factors potentially influencing patient prognosis.
Next, WES and metabolomics data were integrated to identify molecular subtypes using unsupervised clustering. The resulting subtypes were analyzed in terms of metabolic profiles, molecular alterations, clinicopathological characteristics, and prognostic outcomes. By combining genomic and metabolic features, the study established a novel molecular classification system for gastric cancer closely associated with genetic and metabolic alterations. This integrated approach offers new insights into the heterogeneity of gastric cancer and provides a foundation for developing precision therapeutic strategies.